Genomic Data Processing with GenomeFlow
Published in DTMBIO 2022.
Harvard Medical School
Harvard Medical School
Harvard Medical School
Abstract
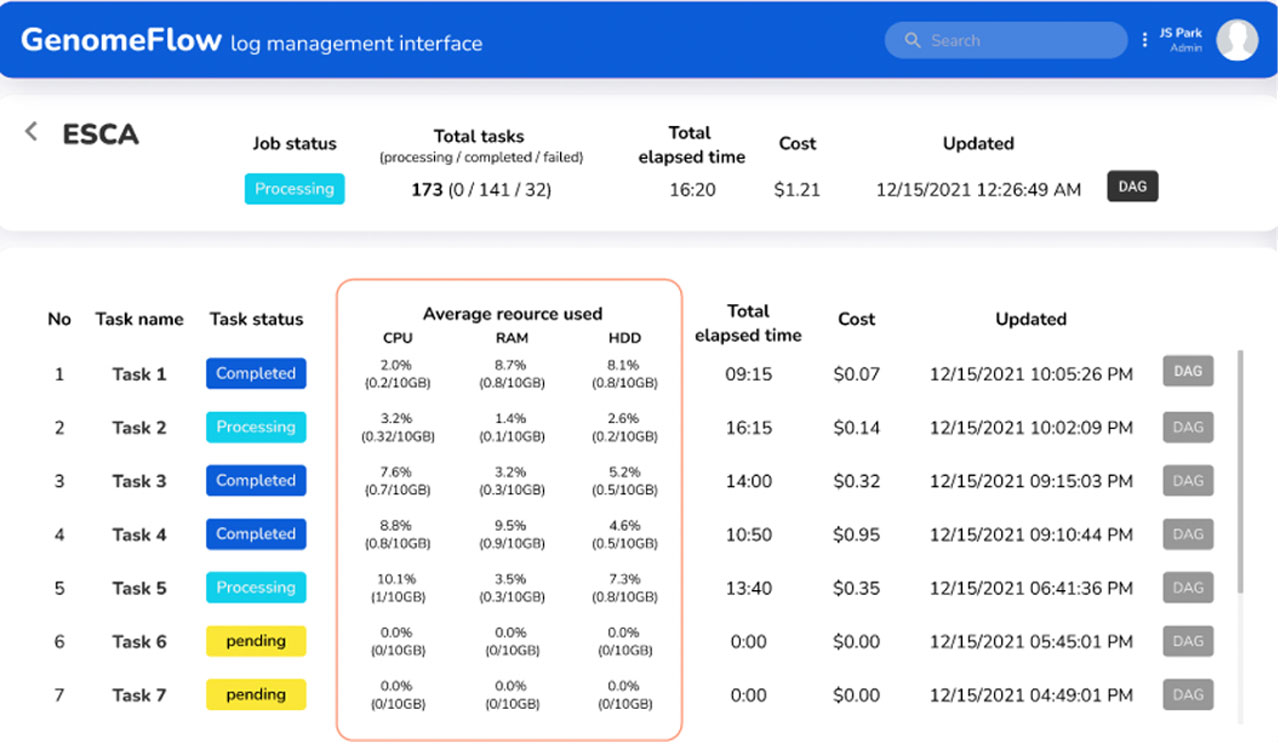
Continuous development of genomics data analysis technologies and expansion of computing storage drive the generation of massive amounts of sequence data, which researchers can share and access through publicly open repositories. On-demand infrastructure services on cloud computing platforms support the processing of such large-scale genomics sequence data in distributed processing environments and can be used to reduce the time of analysis. However, parallel processing methods on cloud computing platforms still present a host of problems for the average user. In particular, cloud computing technology can be difficult to understand when designing an infrastructure suitable for a pipeline, and there is a risk that costs may increase exponentially if computing resources are not properly allocated. To overcome these challenges, we developed an automated infrastructure development and resource optimization program called GenomeFlow, a tool that is able to process large-scale samples at a minimal cost. Here, we describe the step-by-step protocol to use GenomeFlow according to a general sample processing scenario. We introduce the protocol for a bioinformatician with no experience in cloud computing and large data processing, which we estimate will take about 4-5 hours to execute.
Materials